
Professor & Head
Qualification : PhD (Banasthali Univ.), PDF (Univ. of Lausanne, Switzerland)
- asheesh@cusb.ac.in
- ashomics@gmail.com
- :
Specialization :
Sequence Analysis, Evolutionary Structure Analysis, Data Mining, Database and Server Development
Experience : 16 years
(Teaching / Research) :
Dr. Asheesh Shanker received postdoctoral training at University of Lausanne, Switzerland and is actively involved in bioinformatics teaching/research since 2004. He published several research papers in peer-reviewed journals of national & international repute and also authored/co-authored 4 books.
My group works on computational data mining, development of computational pipeline for biological data analysis and database development. Apart from this, I start exploring the role of evolution in drug resistance, its effect on protein structure, and adaptability of a pathogen against its host.


- Publications :
- Priya P, Shanker A (2021) Coevolutionary forces shaping the fitness of SARS-CoV-2 spike glycoprotein against human receptor ACE2. Infection, Genetics and Evolution 87: 104646
- Jayaswal PK, Shanker A, Singh NK (2020) Genome wide annotation and characterization of young, intact long terminal repeat retrotransposons (In-LTR-RTs) of seven legume species. Genetica 148: 253-268.
- Kumar S, Kumari S, Shanker A (2020) In silico mining of simple sequence repeats in mitochondrial genomes of genus Orthotrichum. Journal of Scientific Research 64: 179-182.
- Kumar S, Shanker A (2020) Length variation of chloroplast simple sequence repeats in the genus Eucalyptus L’Hér. Plant Science Today 7(3): 353-359
- Jayaswal PK, Shanker A, Singh NK (2020) Genome-wide comparative and evolutionary analysis of transposable elements in eight different legume plants Indian Journal of Agricultural Sciences 90: 1025-1031.
- Kumar S, Shanker A (2020) In silico comparative analysis of simple sequence repeats in chloroplast genomes of genus Nymphaea. Journal of Scientific Research 64: 186-192.
- Anand K, Kumar S, Alam A, Shanker A (2019) Mining of microsatellites in mitochondrial genomes of order Hypnales (Bryopsida) Plant Science Today;6(sp1): 635-638
- Jayaswal PK, Shanker A, Singh NK (2019) Phylogeny of actin and tubulin gene homologs in diverse eukaryotic species. Indian J Genet 79: 284-291 DOI:10.31742/IJGPB.79S.1.20.
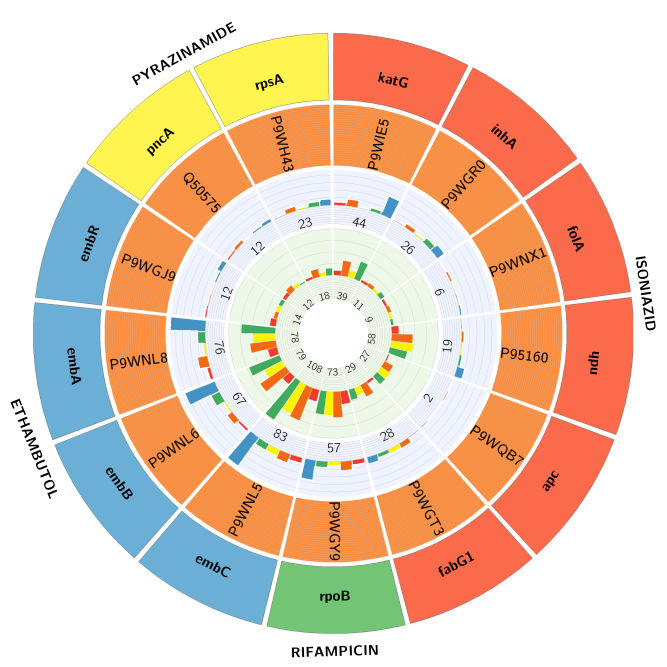
- Vats S, Shanker A (2019) Groups of coevolving positions provide drug resistance to Mycobacterium tuberculosis: a study using targets of first-line antituberculosis drugs. International journal of antimicrobial agents 53: 197-202
- Singh AK, Yadav S, Sharma K, Firdaus Z, Aditi P, Neogi K, Bansal M, Gupta MK, Shanker A, Singh RK, Prakash P (2018) Quantum Curcumin mediated inhibition of gingipains and mixed-biofilm of Porphyromonas gingivalis causing chronic periodontitis. RSC Advances 8: 40426- 445
- Kumar S, Shanker A (2018) Common, unique, and polymorphic simple sequence repeats in chloroplast genomes of genus Arabidopsis. Vegetos 31 (Special): 125-131
- 12. Goyal S, Jamal S, Shanker A, Grover A (2018) Structural basis for drug resistance mechanisms against anaplastic lymphoma kinase. Journal of Cellular Biochemistry 120: 768-777
- Rawat N, Shanker A, Joshi GK (2018) Bioinformatics analysis of a novel Glutaredoxin gene segment from a hot spring metagenomic DNA library. Meta Gene 18: 107-111
- Shukla N, Kuntal H, Shanker A, Sharma SN (2018) Mining and analysis of simple sequence repeats in the chloroplast genomes of genus Vigna. Biotechnology Research and Innovation 2: 9-18
- Shukla N, Kuntal H, Shanker A, Sharma SN (2018) Hydro-priming methods for initiation of metabolic process and synchronization of germination in mung bean (Vigna radiata L.) seeds. Journal of Crop Science and Biotechnology 21: 137-146
- Jamal S, Goyal S, Shanker A, Grover A (2017) Predicting neurological adverse drug reactions based on biological, chemical and phenotypic properties of drugs using machine learning models. Nature Scientific Reports 2017; DOI: 10.1038/s41598-017-00908-z
- Goyal S, Jamal S, Shanker A, Grover A (2017) Structural basis for drug resistance mechanisms against EGFR. Current Topics in Medicinal Chemistry 17: 1-13
- Jamal S, Goyal S, Shanker A, Grover A (2017) Machine learning and molecular dynamics based insights into mode of actions of insulin degrading enzyme modulators. Combinatorial Chemistry and High Throughput Screening 20: 1-13
- Shanker A (2016) Structural Analysis of Form I Ribulose-1, 5-Bisphosphate Carboxylase/Oxygenase BAOJ Bioinfo 1: 1
- Srivastava D, Shanker A (2016) Identification of simple sequence repeats in chloroplast genomes of Magnoliids through bioinformatics approach. Interdisciplinary Sciences: Computational Life Sciences 8: 327-336 DOI 10.1007/s12539-015-0129-4
- Jamal S, Goyal S, Shanker A, Grover A (2016) Computational screening and exploration of disease-associated genes in Alzheimer’s disease. Journal of Cellular Biochemistry 9999: 1-9
- Jamal S, Goyal S, Shanker A, Grover A (2016) Integrating network, sequence and functional features using machine learning approaches towards identification of novel Alzheimer genes. BMC Genomics 17: 807
- Shanker A (2016) Detection of simple sequence repeats in the chloroplast genome of Tetraphis pellucida Hedw. Plant Science Today 3: 207-210
- Kabra R, Kapil A, Attarwala K, Rai PK, Shanker A (2016) Identification of common, unique and polymorphic microsatellites among 73 cyanobacterial genomes. World J Microbiol Biotechnol 32: 71
- Singh A, Kaushik R, Mishra A, Shanker A, Jayaram B (2015) ProTSAV: A protein tertiary structure analysis and validation server. BBA Proteins and Proteomics 1864: 11-19
- Shanker A (2015) An update on sequenced chloroplast genomes of Bryophytes. Plant Science Today 2: 172-174
- Srivastava D, Shanker A (2015) In silico mining and analysis of simple sequence repeats in chloroplast genomes of order Rosales. Research Journal of Bioinformatics 2: 1-9
- Goyal S, Jamal S, Shanker A, Grover A (2015) Structural investigations of T854A mutation in EGFR and identification of novel inhibitors using structure activity relationships. BMC Genomics 16(Suppl 5):S8
- Jamal S, Goyal S, Shanker A, Grover A (2015) Checking the STEP-associated trafficking and internalization of glutamate receptors for reduced cognitive deficits: A machine learning approach-based cheminformatics study and its application for drug repurposing. PlosOne 10: e0129370
- Kapil A, Rai PK, Shanker A (2014) ChloroSSRdb: a repository of perfect and imperfect chloroplastic simple sequence repeats (cpSSRs) of green plants. Database DOI 10.1093/database/bau107 [IF: 2.627]
- Jugran J, Joshi GK, Bhatt JP, Shanker A (2016) Production and partial characterization of extracellular protease from Bacillus sp. GJP2 isolated from a hot spring. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 86: 171. DOI 10.1007/s40011-014-0434-4
- Shanker A (2014) Simple sequence repeats mining using computational approach in chloroplast genome of Marchantia polymorpha. Arctoa 23: 145-149
- Shanker A (2014) Computational mining of microsatellites in the chloroplast genome of Ptilidium pulcherrimum, a liverwort. International Journal of Environment 3: 50-58
- Kumar M, Kapil A, Shanker A (2014) MitoSatPlant: Mitochondrial microsatellites database of viridiplantae. Mitochondrion 19: 334-337. http://dx.doi.org/10.1016/j.mito.2014.02.002
- Shanker A, Sharma V (2012) Efficacy of maximum likelihood method and longer sequences in phylogenetic analysis. Res J Biotech 7: 114-118
- Shanker A (2012) Genome research in the cloud. OMICS A Journal of Integrative Biology 16: 422-428
- Shanker A, Sharma V, Daniell H (2011) Phylogenomic evidence of bryophytes’ monophyly using complete and incomplete data sets from chloroplast proteomes. JPBB 20: 288-292
- 38. Shanker A, Sharma V, Daniell H (2009) A novel index to identify unbiased conservation between proteomes. IJIB 7: 32-38
- Shanker A, Sharma V (2009) Annotation Jargon- It’s not too late to correct it. Current Science 97: 983-984
- Shanker A, Bhargava A, Bajpai R, Singh S, Srivastava S, Sharma V (2007) Bioinformatically mined simple sequence repeats in UniGenes of Citrus sinensis. Scientia Horticulturae 113: 353-361
- Shanker A, Singh A, Sharma V (2007) In silico mining in expressed sequences of Neurospora crassa for identification and abundance of microsatellites. Microbiological Research 162: 250-256
- Shanker A, Tripathi D, Khandelwal G, Sharma G, Agrawal S, Singh A, Sharma V (2006) Data Mining and Database Development for Simple Sequence Repeats of Takifugu rubripes (Japanese Pufferfish) Biochem Cell Arch 6: 1-12
- Satyapal GK, .. , Shanker A, Kumar N (2022) Cloning and functional characterization of arsenite oxidase (aoxB) gene associated with arsenic transformation in Pseudomonas sp. strain AK9. Gene 146926 doi: 10.1016/j.gene.2022.146926
- Kumar S, Singh A, Kumar N, Choudhary M, Choudhary BK, Shanker A (2022) AutomAted RepeaT Identifier (AARTI): A tool to identify common, polymorphic, and unique microsatellites. Mitochondrion 65: 161-165
- Kumar S, Singh A, Shanker A (2022) pSATdb: a database of mitochondrial common, polymorphic, and unique microsatellites. Life Science Alliance 5: e202101307; DOI: 10.26508/lsa.202101307
- Book/Book Chapters :
- Shanker A (2018) Bioinformatics: Sequences, Structures, Phylogeny. Springer Nature Singapore Pte Ltd, p. 402 (ISBN 978-981-13-1561-9)
- Shanker A (2015) The programming language “Perl” for Biologists: Solutions for Beginners. GRIN Verlag GmbH, Germany, p. 188 (ISBN 978-3-656-90770-1)
- Shanker A, Sharma V (2012) Evolutionary analysis of plants using chloroplast proteome sequences. LAP Germany, p. 152 (ISBN 978-3-8473-7563-0)
- Sharma V, Munjal A, Shanker A (2008; 2016 IInd Edition) A Text Book of Bioinformatics. Rastogi Publications, Meerut, India, p. 260 (ISBN 978-81-7133-917-4); IInd Edition p. 350 (ISBN 978-93-5078-142-5)
- Awards :
Indo Swiss Joint Research Programme (ISJRP) fellowship availed at University of Lausanne, Switzerland (2010). - Projects :
Comparison of chloroplast proteomes of bryophytes and pteridophytes to identify orthologous proteins and transition link between both lineages, UGC
Establishment of a server to compare small genomes/proteomes and generate concatenated gene/protein sequences dataset to be used in phylogenetic tree reconstruction, UGC











